Skimming in VIBE#
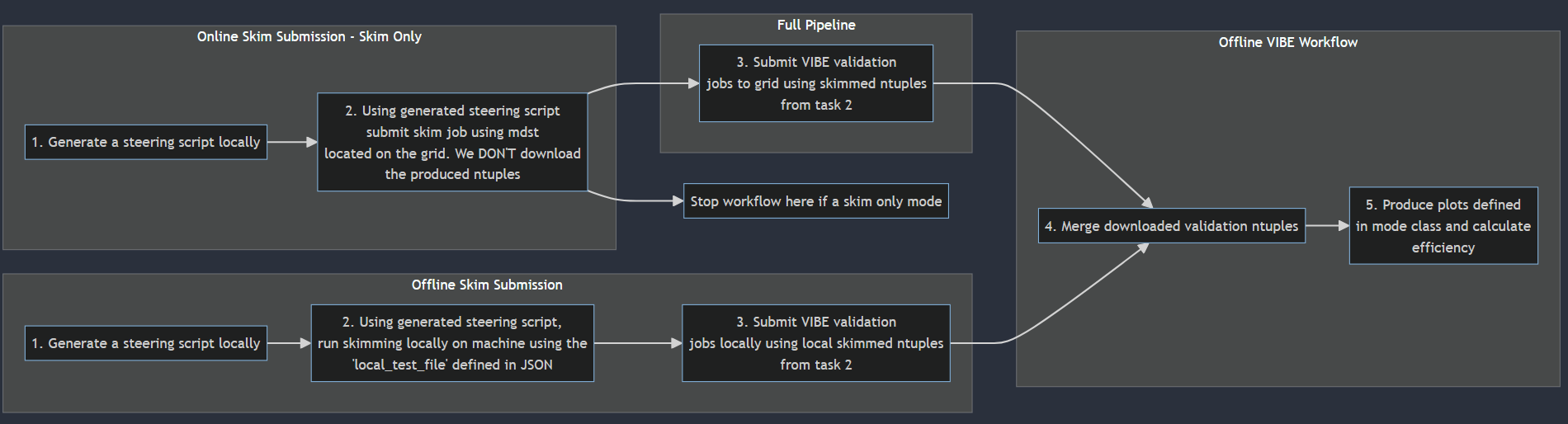
This is a detailed overview of how skimming in VIBE works, what you can do with it and how the operations work.
There are two ways in which you can run skims in VIBE:
Type 1: Skim Only
Will only submit skims to the grid, monitor and reschedule.
Ideal for skimming over any data type using any skim in Belle II on on the grid
Single and Combined skims are available
Use case: Autonamously skim generic or signal MC for Belle II (Email Cameron about the WG1 skimming software for WG specific run dependent skimming and documentation!)
Type 2: Skim and Validation
Run the full VIBE workflow pipeline.
Skims, validates and produces output plots as specified in the skim Mode class
Single and Combined skimming available
Enables any combination of skims and datasets to be validated using VIBE’s framework
Use case: Autonamously skim small sets of generic signal MC and then validate using histograms and statsical metrics
Coming soon: Skimming in VIBE will also be able to calculate retention rates on larger sample sizes than the nightly validation.
Creating the Skim Mode python file#
Analogous to the Creating a Validation Mode in VIBE, a skim mode is defined within a python class derived from ValidationModeBaseClass. To begin however, we need to create a new python script inside one of the subdirectories of vibe/skim_production_modes. If we look inside this directory we see folders for some of the working groups in Belle II along with an example directory:
$ ls vibe/skim_production_modes/
example slme
If your working group is not listed, open an issue on the VIBE github and a directory will be added for your working group along with the necessary changes to the backend to enable VIBEs functionality.
For this tutorial, we will create a new python script inside of the example subdirectory. The naming convention for modes is detailed in Creating a Validation Mode in VIBE:
$ touch vibe/skim_production_modes/example/docSkimExampleMode.py
Skim Mode Template Part 1#
The structure of a skim mode is nearly identical to that described in Creating a Validation Mode in VIBE, except there is one additional required method get_skim_attributes:
# vibe/skim_production_modes/examples/docSkimExampleMode.py
import basf2
from typing import List
from vibe.core.utils.misc import fancy_validation_mode_header
from vibe.core.validation_mode import ValidationModeBaseClass
from vibe.core.helper.skims.skim_attribute_tools import SkimAttributes, AnalysisParameters
from vibe.core.helper.histogram_tools import HistVariable, Histogram, HistComponent
__all__ = [
"DocsSkimExampleMode"
]
@fancy_validation_mode_header
class DocsSkimExampleMode(ValidationModeBaseClass):
name = 'docSkimExampleMode'
def get_skim_attributes(self):
return SkimAttributes(...)
def create_basf2_path(self) -> basf2.Path:
path = basf2.Path()
...
return path
@property
def analysis_validation_histograms(self) -> List[Histogram]:
...
return [hist1, hist2,...]
The get_skim_attributes method returns a SkimAttributes object. This object controls everything about your skim in VIBE. The SkimAttributes object has the following form:
SkimAttributes(
skim_name : List[str],
globaltag : str,
skim_to_analysis_pipeline : Union[bool, AnalysisParameters]
)
Where:
skim_name: List of required skims to run. Either a single element list or can be overloaded to produce a combined skim.
globaltag: The analysis globaltag required for the skim (required for FEI)
skim_to_analysis_pipeline: This toggles between the two different run types of skimming in VIBE. If False, Type 1 will toggle and only the skimming section will run. If True then the full skim to analysis pipeline will run. However, if additional information needs to be passed to the validation portion of the pipeline, the object AnalysisParameters can be passed, allowing the user to add that required information
AnalysisParameters#
AnalysisParameters has two attributes which may be needed by the user for the validation portion of the pipeline
AnalysisParameters(
kwargs : Dict[str,Dict[str,str]]
globaltags : List[str]
)
Where
kwargs: Is a dictionary indexed by the associated skim name inside the skim_name list i.e the key must match one of the given skims. The values of kwargs is a dictionary of keyword arguments to be passed to the create_basf2_path function in your skim Mode. This would usually be added to the associated JSON file alongside the lpn. In skim modes we cannot just add to the JSON file before running VIBE, instead it needs to be taken from the AnalysisParameters and added to the JSON file during run time
globaltags: Any globaltags that need to be prepended to the validation basf2 path.
Skim Mode Template Part 2#
with the lessons learnt in Part 1 we can now fill out our skim mode template.
# vibe/skim_production_modes/examples/docSkimExampleMode.py
import basf2
from typing import List
from vibe.core.utils.misc import fancy_validation_mode_header
from vibe.core.validation_mode import ValidationModeBaseClass
from vibe.core.helper.skims.skim_attribute_tools import SkimAttributes, AnalysisParameters
from vibe.core.helper.histogram_tools import HistVariable, Histogram, HistComponent
__all__ = [
"DocsSkimExampleMode"
]
@fancy_validation_mode_header
class DocsSkimExampleMode(ValidationModeBaseClass):
name = 'docSkimExampleMode'
def get_skim_attributes(self):
return SkimAttributes(
skim_name = ['feiSL', 'feiHadronic'],
globaltag = 'analysis_tools_light-2305-korat',
skim_to_analysis_pipeline = AnalysisParameters(
kwargs= {
"feiSL" : {"treename" : "B0:feiSL"},
"feiHadronic" : {"treename" : "B0:feiHadronic"}}
)
)
def create_basf2_path(self, treename) -> basf2.Path:
path = basf2.Path()
# Truth match using the treename passed into the function
ma.matchMCTruth(
treename,
path=path
)
# -- Do some more analysis stuff
vm.addAlias('sigProbRank', 'extraInfo(sigProbRank)')
vm.addAlias('cosThetaBY', 'cosThetaBetweenParticleAndNominalB')
vm.addAlias("dmID", "extraInfo(decayModeID)")
self.variables_to_validation_ntuple(
decay_str=treename,
variables=list(
{
"cosThetaBY",
"isSignalAcceptMissingNeutrino",
"sigProbRank",
"dmID",
"Mbc",
}
),
path=path,
)
return path
@property
def analysis_validation_histograms(self) -> List[Histogram]:
return [hist1, hist2, ...]
This example shows how AnalysisParameters can be used to pass variables into the create_basf2_path to distinguish between the tree names inside the skimmed rootfiles.
